Main Content
NMR spectroscopy to unravel biomolecular mechanisms
We apply nuclear magnetic resonance (NMR) spectroscopy to resolve the structure and function of proteins and their interactions at atomic resolution and thus unravel their molecular mechanisms.
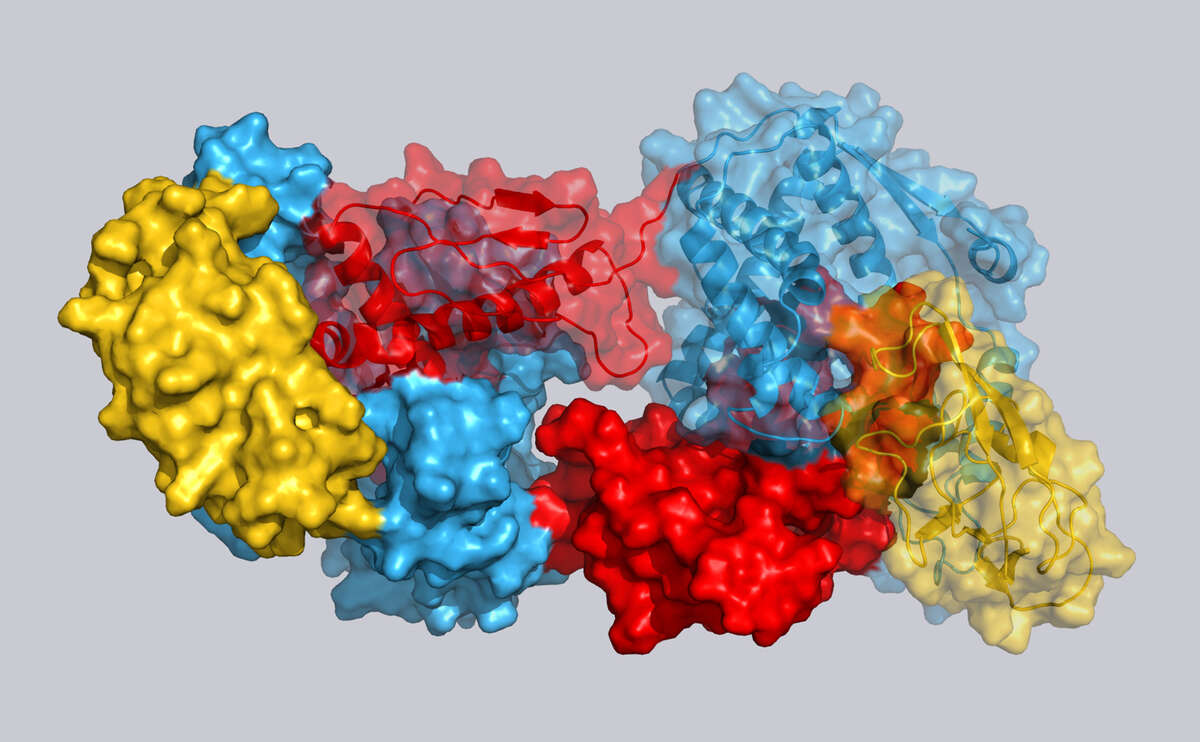
Proteins perform a wide range of essential biological functions, including signal transduction, catalysis, cellular homeostasis, metabolism and membrane transport. We characterize the underlying molecular processes at atomic resolution by solution NMR spectroscopy and related techniques.
Principles of chaperone function
Specific helper proteins, so-called chaperones, ensure the proper formation of other proteins – their clients – in the cell and their transport to the right places. We describe structure and dynamics of chaperone–client complexes at atomic resolution. These descriptions reveal the biophysical laws governing chaperone function and have implications for neurodegenerative diseases, such as Parkinson’s.
Outer membrane protein biogenesis and novel antibiotics
Proteins in the outer membrane of bacteria are produced by an assembly line of molecular chaperones. We investigate the underlying interactions, mechanisms and folding reactions at atomic detail. Of particular interest is the huge insertase BamA that folds and inserts the outer membrane proteins into the membrane. BamA is also a highly valuable target for the novel antibiotics OMPTA and darobactin.
Dynamic kinases and kinase interactions
Kinases govern central cellular functions, including signaling and the regulation of metabolism. We investigate how bacterial kinases are controlled by second messengers and how the mammalian TOR complex interacts with its dynamic substrate proteins to control cell growth and division. These interaction studies between flexible substrates and gigantic kinase molecules benefit optimally from the power of solution NMR spectroscopy.